Imaging plays a crucial role across numerous research areas such as biology, biochemistry, environmental sciences, materials science, and biomedical studies, since it is difficult to understand what we cannot see. Optical imaging stands out as one of the most important imaging modalities due to its non-invasive nature, versatility, and the capacity to visualize phenomena at multiple scales.
My research goal is to catalyze scientific breakthroughs using novel optical imaging techniques.
Nanoscale Imaging Using Single Molecule Orientation Localization Microscopy
One revolutionary advancement in optical imaging that surpasses the diffraction limit is single molecule localization microscopy (SMLM). This technique uses the controlled on and off of fluorescent molecules to image and precisely locate individual molecules. Since it was first proposed nearly two decades ago, microscopists have sought to encode additional parameters associated with single molecules (SMs) to resolve scientific phenomena that cannot be resolved by position alone. A parameter of particular interest is molecular orientation. The imaging technique that captures both the positions and orientations of the molecules is known as single-molecule orientation-localization microscopy (SMOLM).
The primary research focus during my PhD has been the development of SMOLM techniques. An early method we developed is the Tri-spot point spread function (PSF), which uses the non-uniform intensity distribution of the dipole emission pattern of SMs to encode molecular orientation in the captured measurements. Building on my theoretical work in both classical and quantum estimation theory, and inspired by the toroidal dipole emission pattern, we next introduced the radially and azimuthally polarized (raPol) microscope, which effectively separates radially and azimuthally polarized fluorescence, achieving better measurement precision. Notably, this system excels at detecting molecules aligned with the optical axis, as these molecules only emit radially polarized fluorescence. Extending these advancements, we introduced the multi-view reflector (MVR) microscope, which achieves isotropic 3D resolution and a measurement precision that is 1.5 times better for determining 6D SM positions and orientations compared to state-of-the-art techniques.
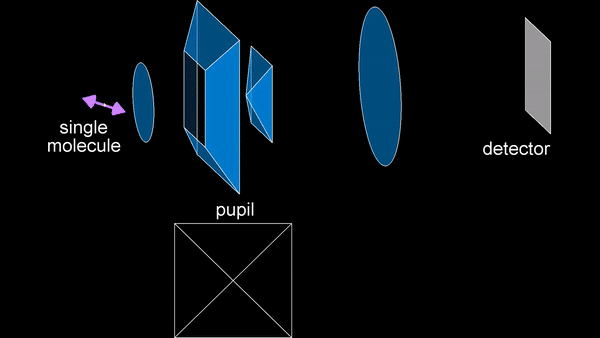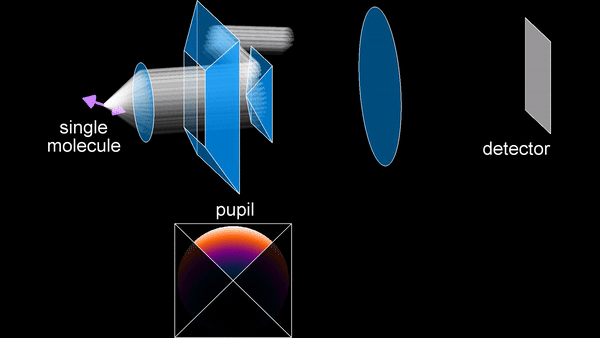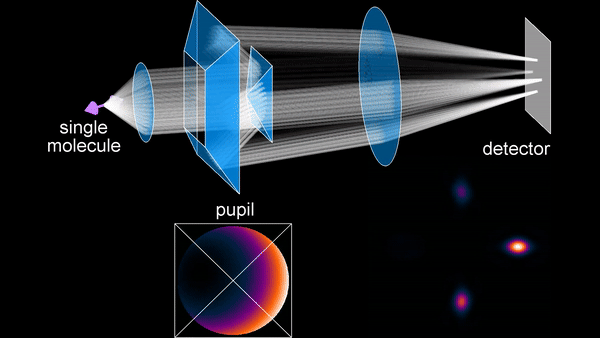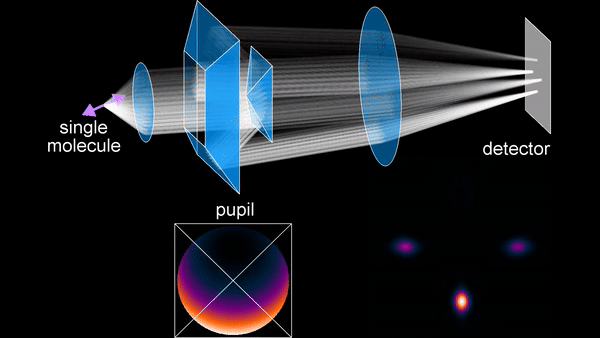
Principle of the MVR microscope.
These methods have enabled the observation of emission anisotropy of fluorescent beads, softening of polymer matrices, compositional heterogeneity in lipid membranes, and infiltration of lipid membranes by amyloid-beta oligomers, which are invisible through conventional SMLM.
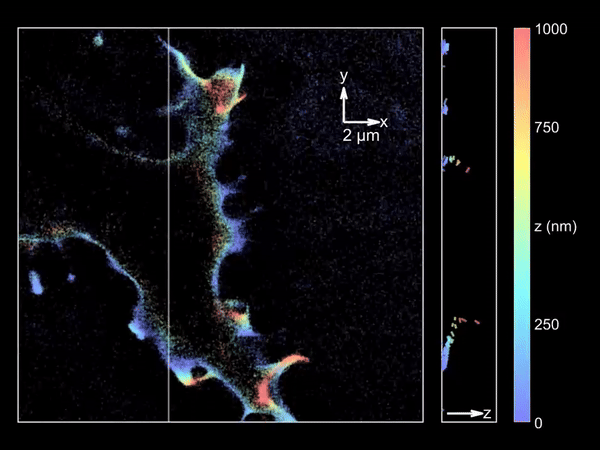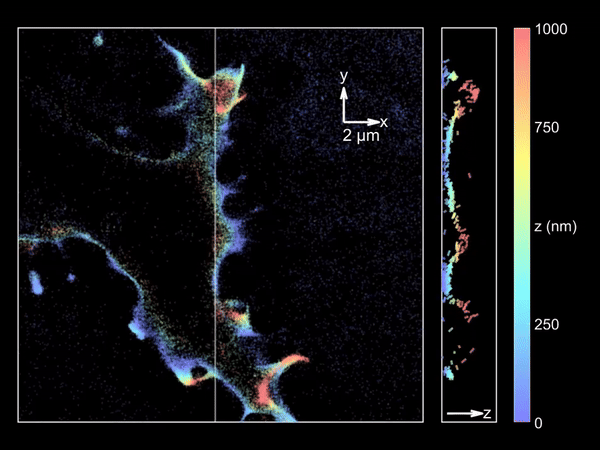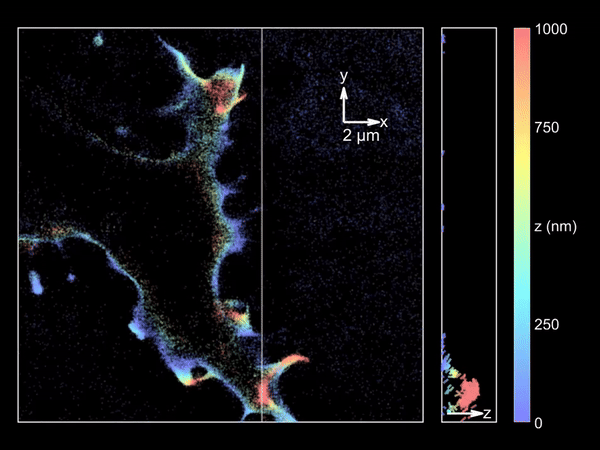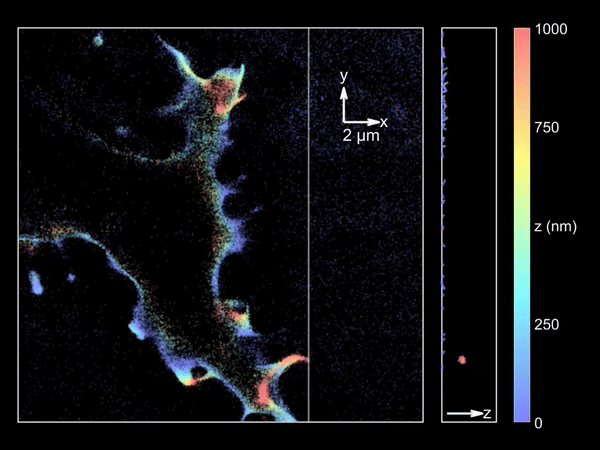
Imaging merocyanine 540 bound transiently to HEK-293T cell membranes.
Computational Imaging for Rhizosphere-Associated Studies
During my postdoctoral research, in collaboration with experts in microbiology, we identified the rhizosphere, the zone of soil surrounding plant roots, as a critical yet underexplored area. Although this region is essential for numerous biological processes, advanced optical imaging techniques are rarely applied to rhizosphere-associated studies. To address this gap, we have developed imaging solutions specifically tailored to these tasks.
The primary challenge in rhizosphere imaging stems from the difficulty of finding a single imaging modality suitable for this complex system. For instance, while fluorescence imaging provides exceptional specificity for differentiating components within the rhizosphere, labeling is not always feasible for environmental samples. This limitation inspired the development of Complex-field and Fluorescence microscopy using the Aperture Scanning Technique (CFAST), a bimodal imaging system that combines 3D fluorescence imaging using an engineered PSF with quantitative phase imaging through the Kramers-Kronig relations. Its bimodal capability has led to significant scientific discoveries, such as tracking of early bacterial aggregate development, bacterial competition, and gene expression under varying environmental conditions.

CFAST imaging of gene expression of P. syn PphoA-mNG under phosphorus limitation. The growth is tracked by the complex field and gene expression by fluorescence.
Another significant challenge in rhizosphere imaging is the necessity for a large field of view given the size of plant roots. High-resolution 3D imaging that requires axial scanning over such expansive areas can be time-consuming. We address this limitation using single-shot volumetric fluorescence (SVF) imaging, which typically encodes 3D data into a single 2D image, followed by computational reconstruction. However, unlike traditional SVF methods, our QuadraPol PSF maps 3D information directly onto a 3D measurement using a polarization camera, overcoming the sparsity constraints. We also developed a reconstruction algorithm with improved reconstruction quality based on neural fields. We have demonstrated the effectiveness of both our hardware and algorithm by imaging bacterial colonies and plant roots. Check out the project webpage here!
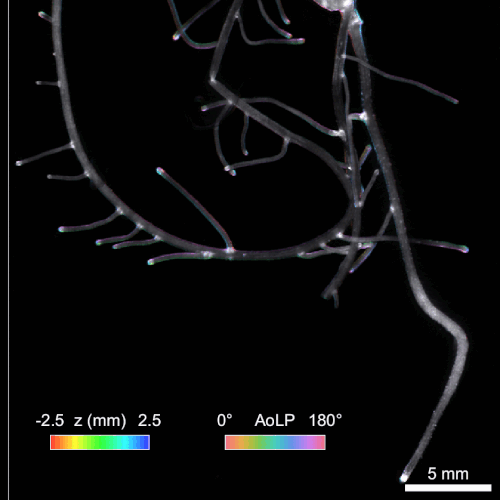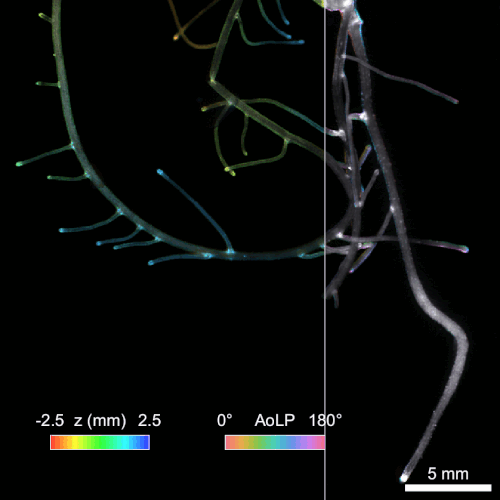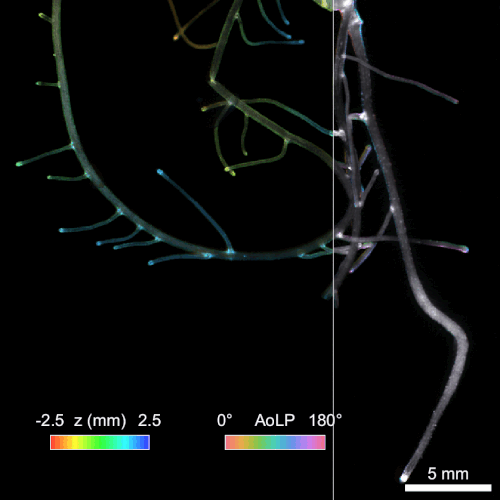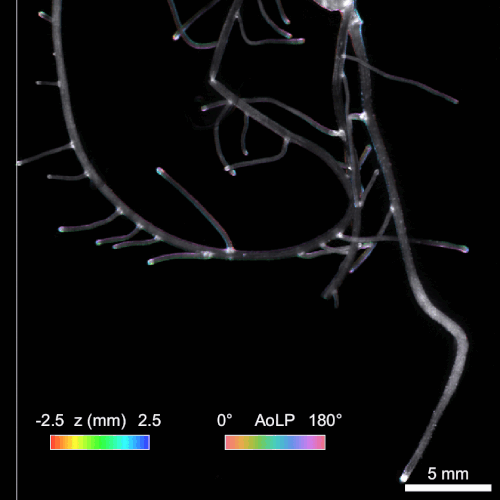
Reconstructing 3D wheat roots using a raw polarized image. AoLP: angle of linear polarization.
